Abstract
Introduction
Although CCAAT/enhancer binding protein alpha double mutated (CEBPADM) acute myeloid leukemia (AML) is considered a low-risk form of AML according to 2017 ELN recommendations, relapse remains a major cause of death. To assess the broader prognostic impact of other cancer-associated genes, we sequenced a panel of 40 myeloid disorders-related genes in a 25 patient cohort.
Methods
16 CEBPADM AML diagnosis samples along with 9 CEBPAsingle mutated (SM) were analyzed by targeted next-generation sequencing (Ion Torrent) using Oncomine Myeloid Research Assay. 4 CEBPADM and 2 CEBPASM AML relapse samples were analyzed as well. All patients received intensive chemotherapy according to 2017 ELN recommendations.
Results
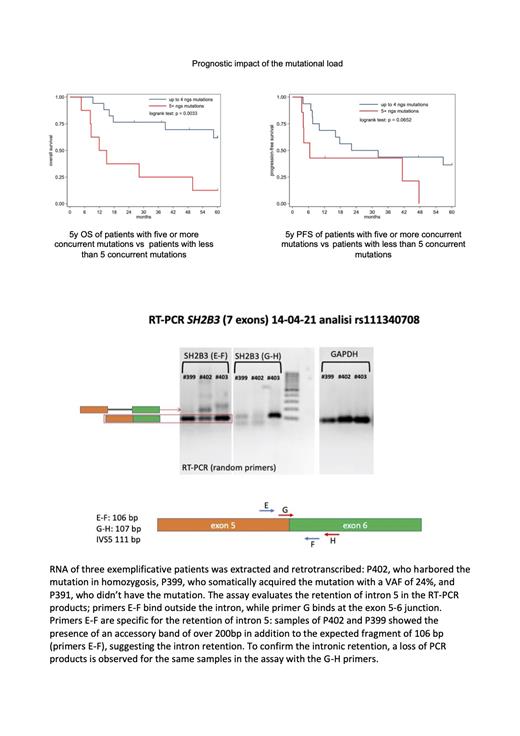
With a median follow-up of 3.2 years (range 0.4-12) 5y OS was 61% and 14% for CEBPADM and CEBPASM patients respectively. Overall, a median of 3 concurrent mutations were present at diagnosis, slightly more in CEBPA SM patients (4 vs 3 in CEBPA DMpatients). The number of somatic mutations influenced both PFS and OS (p = 0.04 and p < 0.01 respectively) independently of CEBPA mutational status. Each single unitary increase in the number of mutations increased the hazard for death of 27.7% (95% CI: -1.4-+65; p = 0.064) while passing from 4 to 5 mutations increased the odds of death by 367%. 5y OS in patients with 5 or more concurrent mutation was 14.3% vs 61.8% in patients with less than 5 co-mutated genes; 5y PFS was 0% vs 38.6%.
Mutational landscape of CEBPADMand CEBPASMAML differed significantly, with GATA2, FLT3, DNMT3A and TET2 being the most frequently mutated genes in CEBPADM vs NPM1, FLT3, DNMT3A and WT1 in CEBPASM patients. NPM1(77.8% vs 6.7%; p < 0.01) and ASXL1 mutations (44.4% vs 0%; p = 0.02) were more frequent in CEBPASM patients, confirming they are mutually exclusive with CEBPA biallelic lesions. DNA methylation was the most frequently mutated pathway in biallelic patients (87%) while chromatin/cohesin complex (88%) was the most frequently mutated one in CEBPA monoallelic patients. Mutations of CEBPA, NPM1, DNMT3A, WT1, STAG2, TET2, ASXL1, IDH2, SRSF2, CALR, PRPF8, NF1, TP53, RUNX1 had the highest median variant allele frequency (VAF), more often representing founding mutations. GATA2, IDH1, KRAS, BCOR, MPL, IKZ2F1 and PTPN11 had a more borderline median VAF, variably being clonal or subclonal. Mutations in the 3 tyrosine kinases genes FLT3-ITD , CSF3R, NRAS were only subclonal.
Mutations in WT1 and FLT3 were associated with increased relapse rate (p = 0.02 and p = 0.01 respectively), while patients with GATA2 mutations had a strong trend towards better 5y OS (83% vs 32%, p = 0.053).
We also identified a not previously described allelic variant in the SH2B3 gene (ATGGGG/A INDEL) with an overall prevalence in our population of 58.3% (46.7% of CEBPA DM and 77.8% of CEBPA SMpatients). Patients with the SH2B3allelic variant had a significantly lower bone marrow blast percentage at diagnosis (p = 0.014) and a strong trend towards a higher number of concurrent mutations (p = 0.056). Moreover, when present at diagnosis, SH2B3 variant persisted at relapse with the same VAF. By real time PCR we demonstrated that this SH2B3 allelic variant leads to a dramatic reduction of the corresponding transcript. This gene encodes for a negative regulator of many crucial signaling pathways (SCF/c-KIT, erythropoietin/JAK2, thrombopoietin/MPL WT/ W515L, JAK2 WT/ V617F, GM-CSFR and FLT3-WT/ITD) of the hematopoietic stem cell.
Matched diagnosis and relapse samples analysis suggested different features of clonal evolution: while mutations of SH2B3, WT1, DNMT3A, NPM1, and IDH1 consistently persisted at relapse, CEBPA and GATA2 mutations were unstable during disease course. ZRSR2 and PRPF8 mutations were found in relapse samples only.
Summary
Our study offers insights into the genetic landscape of CEBPADM mutated AML as compared to CEBPASM AML, highlighting the contribution of NGS to risk stratification. In fact, our data show that the number and the type of concurrent mutation has a prognostic impact, possibly identifying patients eligible to first line allogeneic stem cell transplantation. We identified an allelic variant of SH2B3 that had never been functionally characterized nor associated with AML and that could represent a marker for genetic instability and a potential new target in AML treatment strategies.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal